Proper embryo selection just landed
Herasight is the new kid on the block
Today is an exciting day. First, the HHS announced a more open data policy (in the direction of my suggestions), and second and more importantly, a new company just went public with their embryo selection service. Herasight is the name, which presumably relates to Hera, the Greek goddess of family and women, which shares the root with heritability. The name thus means ability to see genes, very fitting. Most importantly, they published their validation white paper the same day as well as a blogpost summary, and a small embryo selection simulator on their website. If you just want the summary, read Alex Young's X thread:
I've read the technical paper and blogpost, so you don't have to (but you should). The gist of the papers is as follows:
Embryo selection involves choosing between genetic siblings, and this requires special care in the methodology due to 1) reduced validity of some polygenic scores within families, and 2) reduced genetic variation which makes every polygenic score worse in practice.
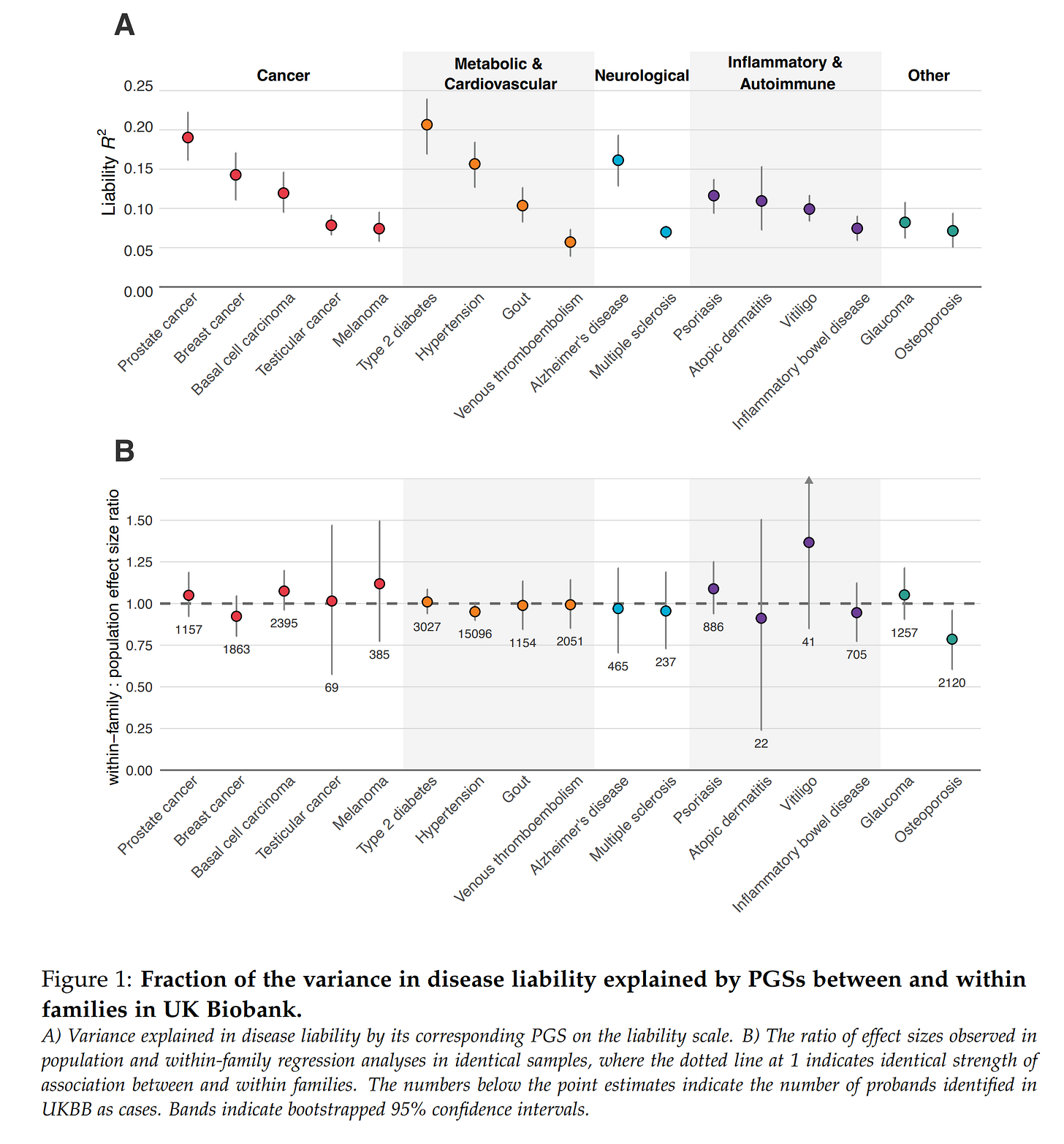
They conducted a comprehensive validation study using unrelated and sibling subjects to estimate model validity and within family declines.
They compared their results to academic results and commercial companies' claims. In the case of Nucleus, it was particularly gruesome as their customer reports present impossible results.
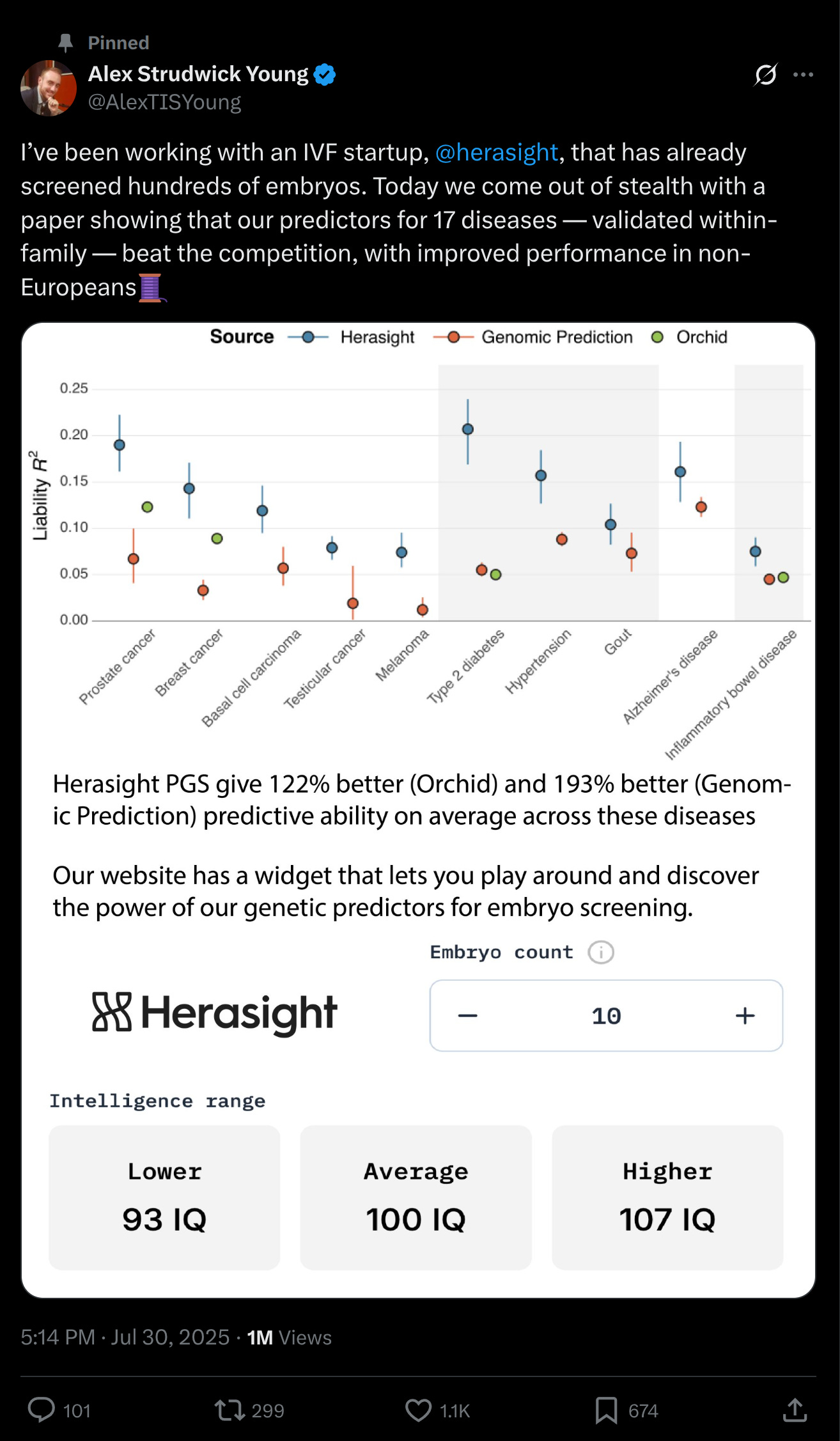
Let's start with the polygenic score validities:
The various models have different levels of validity. This reflects three factors:
Training sample size (rare traits give you little training data but often have simpler genetic architectures)
Inherent difficulty related to genetic architecture, which encompasses polygenicity (more variants of smaller effects are harder to find and estimate precisely) and non-additive or interaction effects (e.g. for immune-system related disorders).
Heritability (you can't produce a good genetic prediction is variation in the phenotype is not much due to genetics).
Given these varying factors, it is nice that they were still able to get to ~20% liability R2 for some of them and none of them really showed any within-family decline (one shows a slight decline, but the confidence interval is very close and there seems to be no multiple testing correction applied). What does this mean in practice? Well, this is where we can use their simulator to get an idea of what a typical couple can expect:
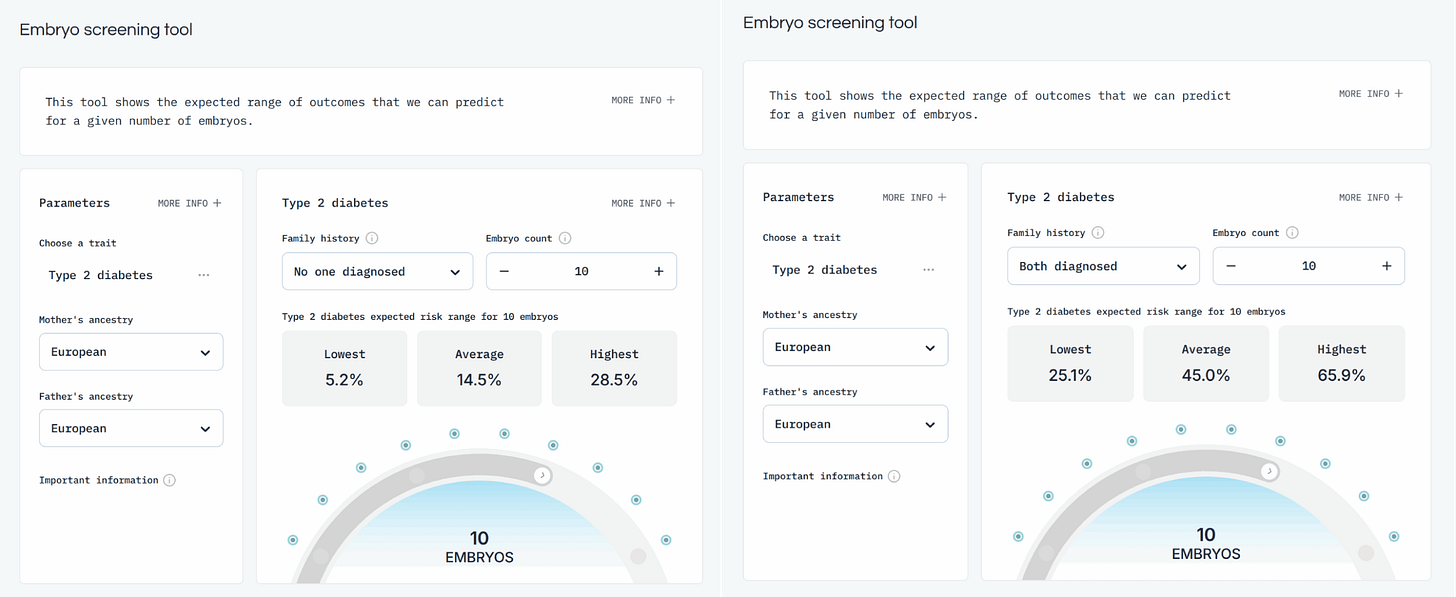
Type 2 diabetes is relatively common, as it is both a disease of old age and of prosperity obesity, both of which are unfortunately common in the Western world. A typical couple may have about 10 euploid (correct number of chromosomes) embryos to choose from. Given this scenario, on average, the worst embryo (for this phenotype) will have a risk of ~29% and the best a risk of only ~5%, roughly a 6x difference. If, on the other hand, your parents are both diabetic, the family-related risk is starkly increased from ~15% to 45%. The worst embryo will on average have a risk of ~66% and the best ~25%, only a ratio of ~2.6. The relative ratio is, however, not what many would be parents (should) care about, but the absolute risk reduction. Using the service, they could roughly reduce the risk of diabetes by ~40%points, which is a massive difference. It doesn't quite make the family-related risk go away (~25 vs. ~15%), but it is approaching that. This is what a polygenic score of ~20% liability R2 can give you. Not perfect, but good, and probably worth it in many cases. Of course, given current trends regarding effective obesity and thus diabetes treatment (Ozempic like GLP-1 drugs), this phenotype may take less priority.
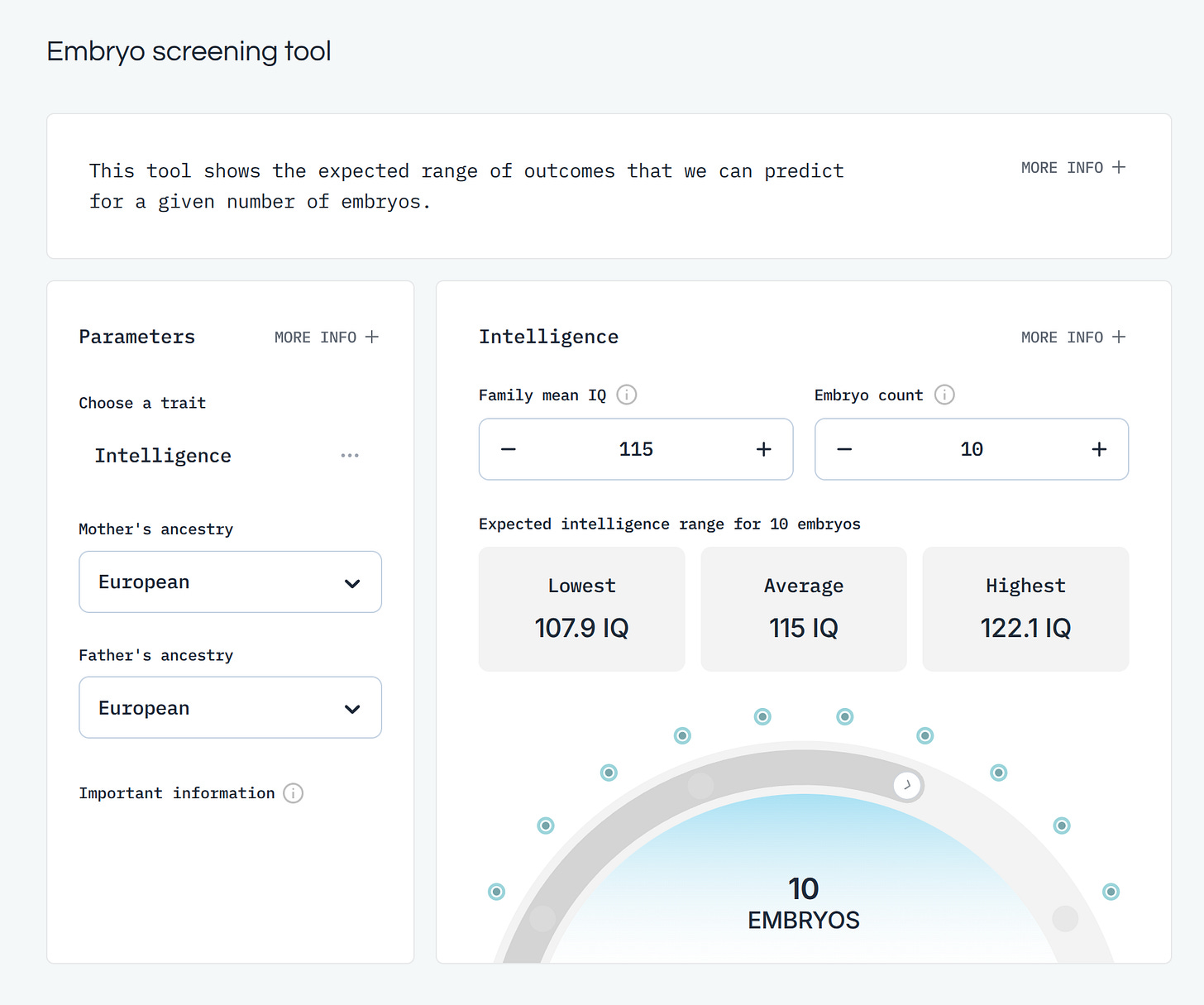
In a similar vein, we can look at their intelligence (IQ) predictor:
In this case, you must input an estimate of the parental (additive) genetic mean IQ. Basically, this is the question of whether you come from 'good stock' intelligence wise. A typical reader of this blog appears to have an IQ around 130 which would statistically mean (assortative mating of ~0.5) the other spouse is around 115 IQ, for a mean phenotypic IQ of 123. Given some regression towards the mean, the parental mean genotypic IQ is about 115 (that is, assuming additive heritability of 70% from 123 IQ). Picking the smartest embryo will (on average!) result in 122 IQ, or a gain of 7 IQ relative to expectations (not using embryo selection), which is still a decline of 1 IQ compared to the parents. The reason for this is that non-perfect heritability fundamentally limits how fast selection can affect a given trait. It does not mean selection is impossible because in the next generation the genotypic mean IQ will have changed upwards (many people mistakenly think regression towards the mean happens each generation and thus selection is impossible because they fail to account for the increase in the genotypic mean trait value).
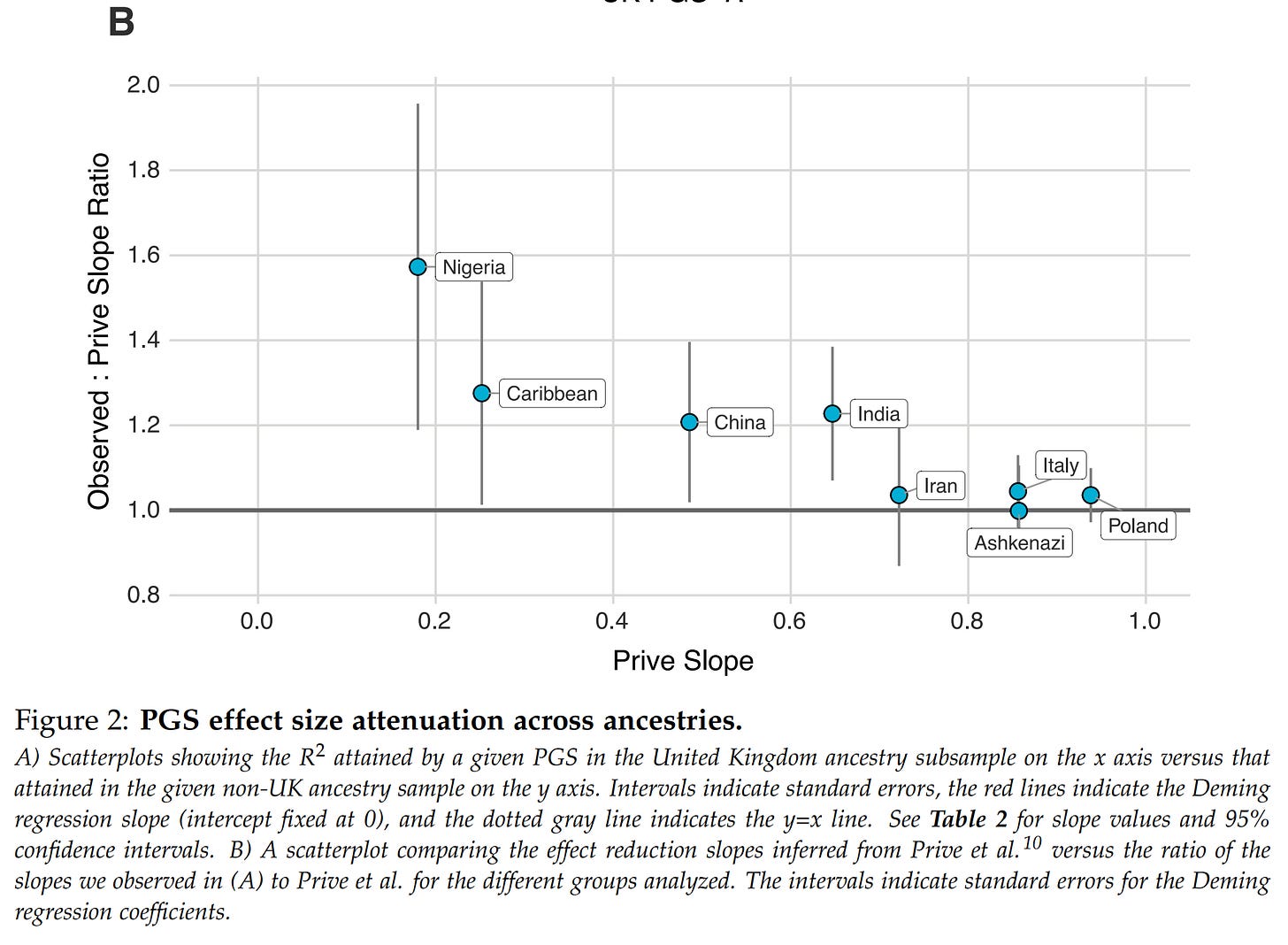
These results concern parents of purely European origin, and specifically, British origin. But most people have some other ancestry, say, Polish or Italian, and some are non-European. How does this affect things? Well, negatively:
Genetic models would ideally find causal variants in the genome (which we could edit directly for much larger improvements), but in general, they cannot, so they find non-causal (tag) variants that are highly correlated with the causal variant(s). These correlations among variants (called LD) differ by population in value (e.g., may be 0.9 in British but 0.8 in Polish), and overall strength. The more inbred a group is (smaller effective population size), the stronger the correlations. Due to these factors, the genetic models using, in part at least, tag variants which will not function as well in other populations because they are specifically chosen for having strong correlations in that population with the causal variant(s). This results in a predictable decline in model validity as a function of genetic distance (this is already known from animal breeding). They validated this expectation on their data, with the expected results that non-British Europeans show minimal less of validity, while Africans show the highest. This makes sense since Africans are both the furthest apart genetically from the British and they have weaker LD (because they are the founding population of humanity, and the others have gone through bottlenecks). This reduction is important because it means the effectiveness of the predictions must be taken into account in the results shown to customers.
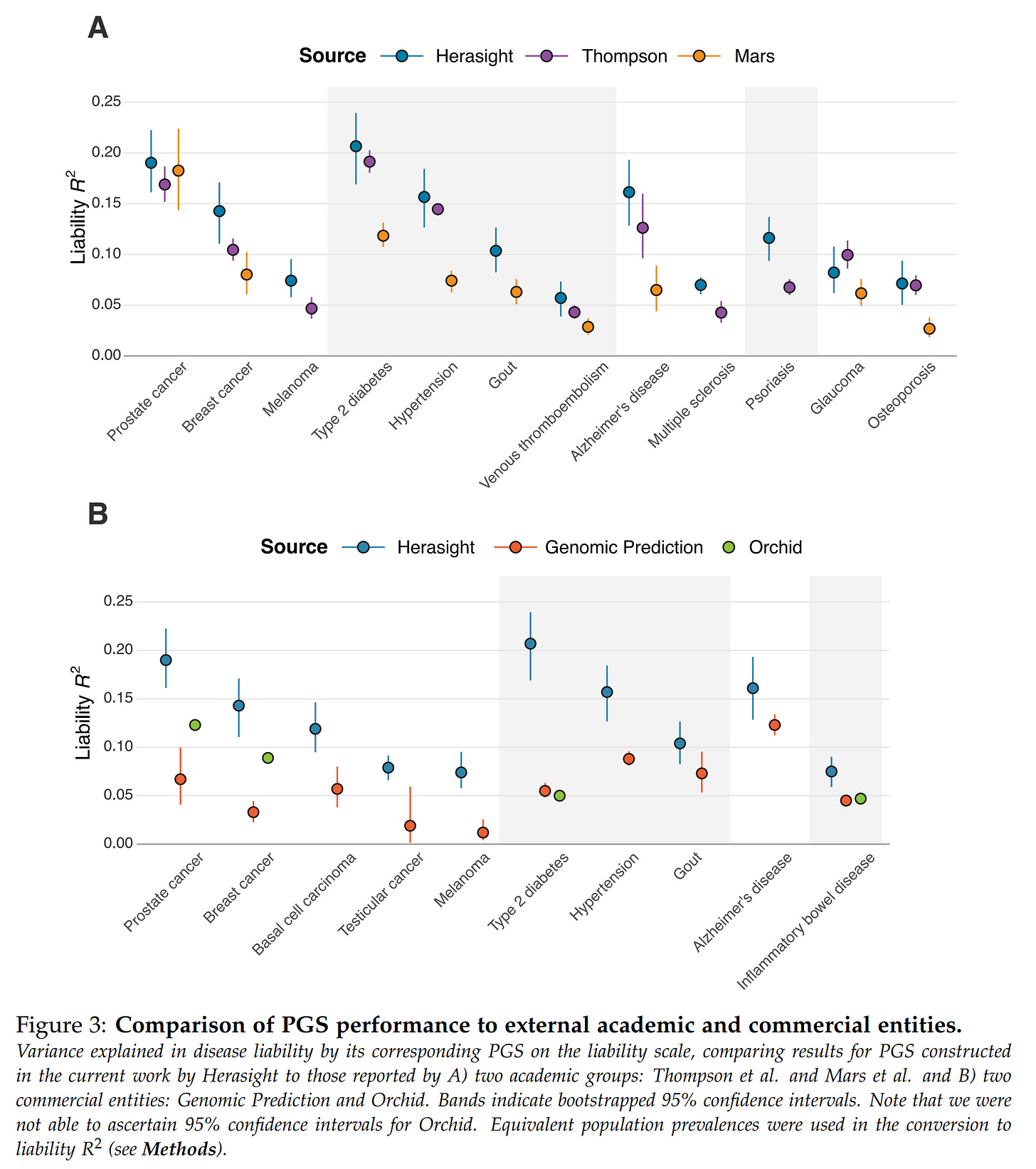
Next up, they analyzed the predictive validities of their custom genetic models versus academic models and commercial competitors:
In general, they do slightly better than the academics, and a lot better than the competitors. I can believe this because the competitors lack researchers with skill in this area and often use outdated off-the-shelf models. It goes to show the value of keeping up to date with the latest GWAS preprints, and doing your own post-GWAS optimization is very beneficial.
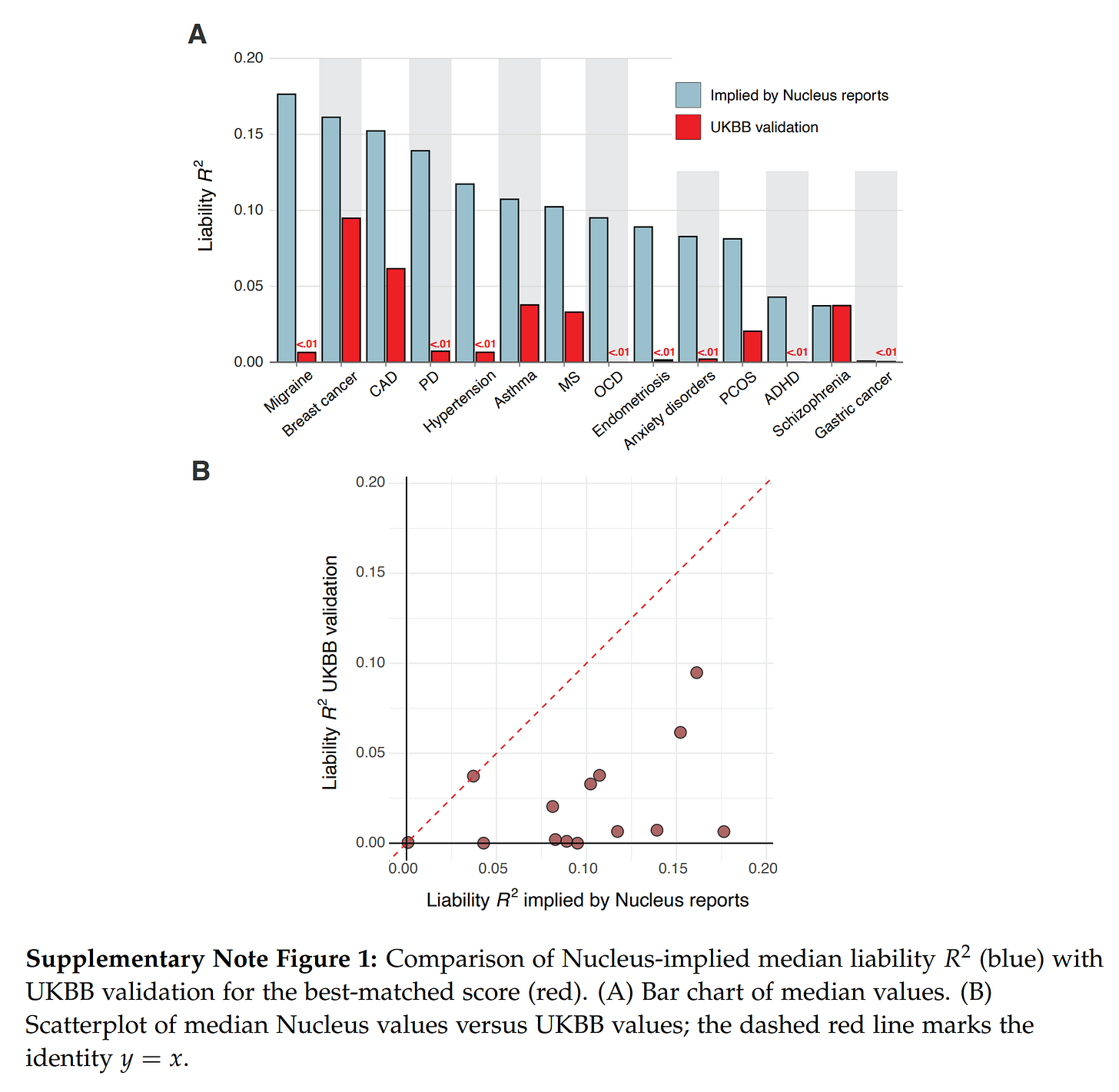
You may also notice that one company is missing here, Nucleus Embryo, why is that? Well, because their investigations of Nucleus reports showed that they contain impossibly good results and some straight up impossible values:
Nucleus gives their customers predictions for embryos like those in the simulator above, but they seem ... too good. Herasight took advantage of the fact that these reported probabilities are convertible to liability R2s if one makes assumptions about the prevalences (e.g. diabetes is 15%). Doing this math shows that the models have implausibly high validities, far beyond any competitors or academic results. Using the source code of the forgone impute.me website (which is archived on certain Github mirrors), it is possible to check whether their current models are the same as those from years ago. Some of these models are downright shoddy, based on only ~12 SNPs! They find that this is mostly the case, meaning that Nucleus has done very little additional R&D before public. And they did this with bad models, but then incorrectly specified them, making them seem much more accurate than they are. In the end, this misleads customers into making wrong decisions (you think you are reducing breast cancer by 20%points, but in reality, it may be 5%points and you should have picked a different embryo). Hopefully, few people have used their service as it is at this time. They have a huge budget (raised >30 million USD!). There are further impossibilities, such as a customer report showing an embryo an elevated polygenic score for ADHD, but somehow has a below average estimated risk. This is just mathematically impossible. Due to these issues, it would make no sense comparing the implicitly claimed validities by Nucleus with the more reliable estimates from Genomic Prediction, Orchid, and Herasight.
Overall, this is great work!


Embryo selection is the killing of a human being. It is highly immoral. It will hasten the demise of what's left of Christendom, which always was the West. God is not mocked.
Great news. It is always good to learn of advancements to improve the human genome.