Species genetic variation and number of subspecies/races/breeds/breeding populations/clusters etc.
A straightforward research idea: Compare the recognized count of subspecies (races/breeds/breeding populations/clusters etc.) with measures of genetic variation. The most obvious is the Fst but it's not an optimal metric. There is some literature on this:
Woodley, M. A. (2010). Is Homo sapiens polytypic? Human taxonomic diversity and its implications. Medical hypotheses, 74(1), 195-201.

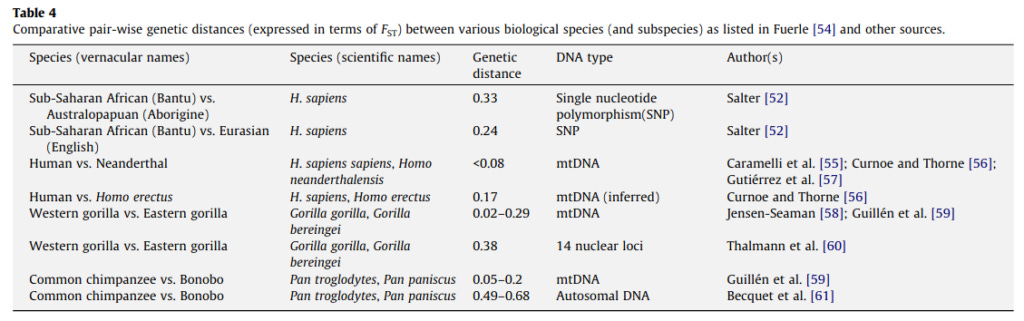
Fuerst, J. (2015). The Nature of Race: The Genealogy of the Concept and the Biological Construct’s Contemporaneous Utility. Open Behavioral Genetics.
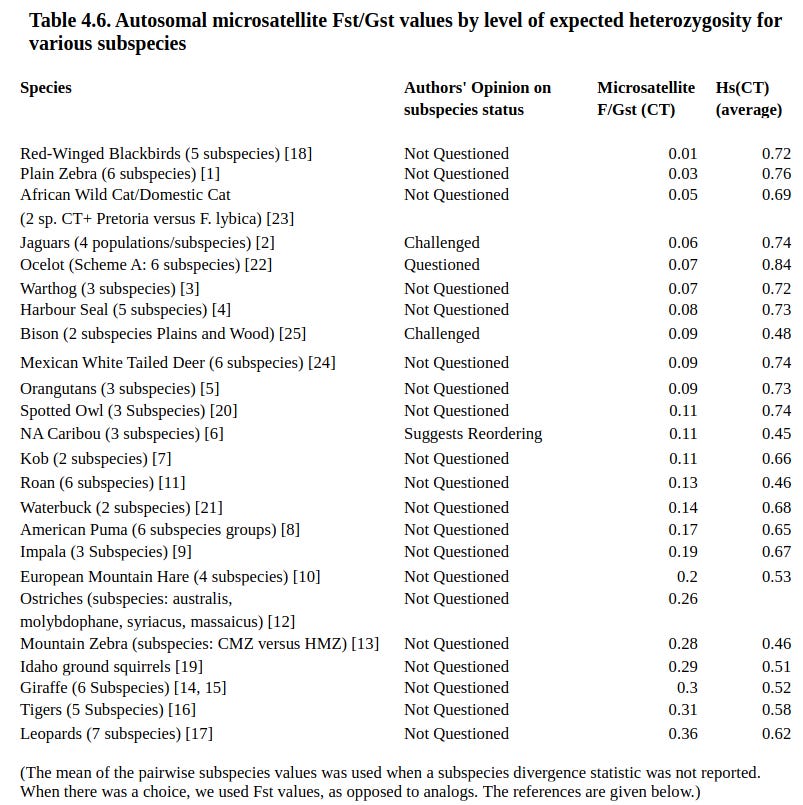

But surely there are newer data since these reviews. One paper I saw a few years ago:
Stoeckle, M. Y., & Thaler, D. S. (2014). DNA barcoding works in practice but not in (neutral) theory. PloS one, 9(7).
This paper has the data.
Thaler, D. S., & Stoeckle, M. Y. (2016). Bridging two scholarly islands enriches both: COI DNA barcodes for species identification versus human mitochondrial variation for the study of migrations and pathologies. Ecology and evolution, 6(19), 6824-6835.
Prior study by authors. Important note "Species without geographically structured or hybrid clusters"
Stoeckle, M. Y., & Thaler, D. S. (2018). Why should mitochondria define species?. BioRxiv, 276717.
More than a decade of DNA barcoding encompassing about five million specimens covering 100,000 animal species supports the generalization that mitochondrial DNA clusters largely overlap with species as defined by domain experts. Most barcode clustering reflects synonymous substitutions. What evolutionary mechanisms account for synonymous clusters being largely coincident with species? The answer depends on whether variants are phenotypically neutral. To the degree that variants are selectable, purifying selection limits variation within species and neighboring species may have distinct adaptive peaks. Phenotypically neutral variants are only subject to demographic processes—drift, lineage sorting, genetic hitchhiking, and bottlenecks. The evolution of modern humans has been studied from several disciplines with detail unique among animal species. Mitochondrial barcodes provide a commensurable way to compare modern humans to other animal species. Barcode variation in the modern human population is quantitatively similar to that within other animal species. Several convergent lines of evidence show that mitochondrial diversity in modern humans follows from sequence uniformity followed by the accumulation of largely neutral diversity during a population expansion that began approximately 100,000 years ago. A straightforward hypothesis is that the extant populations of almost all animal species have arrived at a similar result consequent to a similar process of expansion from mitochondrial uniformity within the last one to several hundred thousand years.


Since they looked at species without clusters, one would need a different dataset. But it should be possible to find this somewhere.

